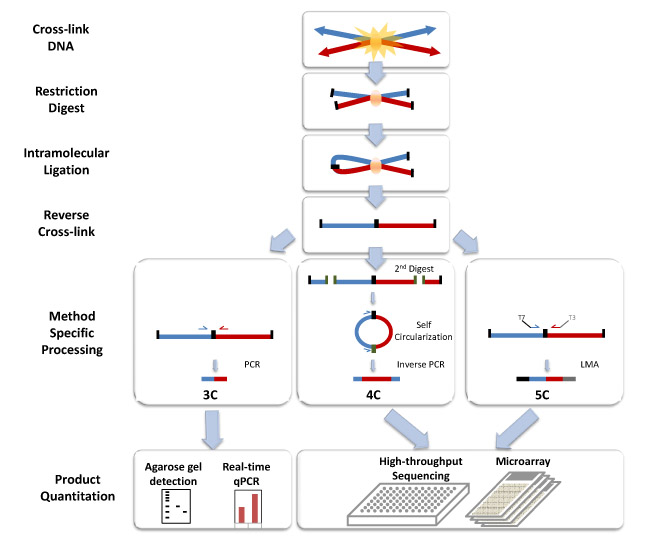
 Since Job Dekker introduced the chromosome conformation capture (3C) approach in 2002, there have been numerous modifications and enhancements to the core method that continue to improve upon its robustness and provide deep insights into one of the biggest unchartered areas of gene regulation.
Since Job Dekker introduced the chromosome conformation capture (3C) approach in 2002, there have been numerous modifications and enhancements to the core method that continue to improve upon its robustness and provide deep insights into one of the biggest unchartered areas of gene regulation.
A decade later, researchers continue to deploy 3C-based methods in a broad range of research settings, thirsty for insights into the elusive and dynamic nature of chromosome structure in normal, disease, and developmental states.
Recently, we pulled data on the majority of “interaction sequencing” methods. These methods include:
-
Protein-DNA interactions (e.g. ChIP Seq)
-
Protein-RNA interactions e.g. CLIP Seq)
-
Chromatin interactions (e.g. 3C)
Global Research Trends in Contact Mapping
Let’s take a closer look at the core applications and methods for contact mapping and how they’ve been used over the years in gene regulation research.
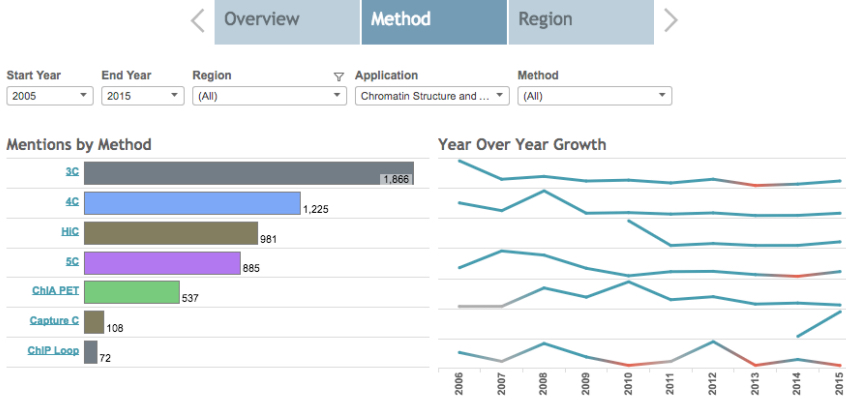
Despite a few rocky years, the core 3C-methods have continued to grow in uptake in labs around the world, which is impressive considering the methods aren’t for the faint of heart. The protocols are fairly complex and data analysis can be challenging. Moreover, few technology firms to come to the aid of researchers in this arena with the exception of TaqMan® 3C Chromosome Conformation Kits that were in early access testing a few years ago, but we’re not sure if they’re available.
This is a screenshot of our interactive Market Trends Dashboard. You can see how the derivative methods have been adopted quickly, but the original 3C approach continues to grow as well (full data and interactivity are available in the Dashboard. Understanding trends are great, but sometimes it’s even more interesting to dive down into the specific institutions and researchers that are behind these trends. That’s where the Research Profile comes in….
Research Profile Dashboard: Top Authors and Universities Using Contact Mapping
We developed the Research Profile Dashboard to provide our clients with a precise view into researcher behavior and activity. As an example below, we show the most active and cited researchers using contact mapping methods, but the same approach can be taken for any:
-
Research Area (e.g. iPSCs, reprogramming, prostate cancer)
-
Application ( e.g. high-throughput sequencing, genome editing, transfection)
-
Method (e.g. ChIP Sequencing, One-Step PCR)
-
Corporate or Product Brand (e.g. Illumina, SuperScript®, FuGENE®